The concept of FoodGuard. A case study
- Designature Community
- Dec 12, 2024
- 1 min read
Updated: Jan 23
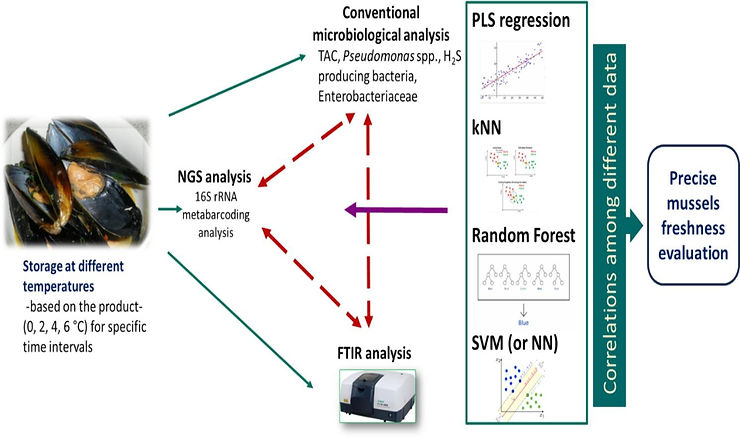
The EU project FoodGuard, is dealing with an enormous amount of heterogeneous data (i.e., microbiological, spectral and Next Generation Sequencing data) obtained from different food being stored under various storage conditions. More specifically, spectral data were collected through Fourier transform Infrared (FTIR) spectroscopy, while the overall profile of microorganisms present in these tested food samples affecting quality and safety, was determined through Next Generation Sequencing (NGS) using 16S rRNA metabarcoding analysis. In parallel, conventional microbiological analysis for the estimation of culturable spoilage microorganisms (total aerobes, Pseudomonas spp., B. thermosphacta, Shewanella spp. and Enterobacteriaceae) was applied. Different machine learning algorithms, namely Partial Least Square (PLS), Support Vector Machines (SVM), k-Nearest Neighbors (kNN), Random Forest (RF) Neural Networks (NN)) were applied accordingly, to assess the potential of FTIR and NGS data to provide useful information about mussels’ microbiological quality.
In particular, the analysis of FTIR data, combined with microbiological and NGS data, revealed spectral regions correlated with mussels’ freshness, including lipid, free fatty acids, protein, peptides, and amino acid changes. It was shown that the application of “multi-omics” in seafood supply chain can provide insightful information about mussels’ quality and safety compared to the methodologies followed in current quality and safety management systems.
More information can be found in our publication in Food Research International 197, 115207.